1. The original intention of MloDisDB
Cells are compartmentalized by numerous membrane-bounded organelles and membraneless organelles (MLOs) to ensure temporal and spatial regulation of various biological processes. Recently, emerging evidence supports that liquid-liquid phase separation (LLPS) is underlying the formation of MLOs, and dysfunction of MLOs are associated with various pathological processes. Despite growing interest in the field, there is a lack of centralized resources presenting relations of MLOs, LLPS and diseases. Herein we build MloDisDB, a curated database which aims to gather MLOs and LLPS related diseases from the dispersed literature.
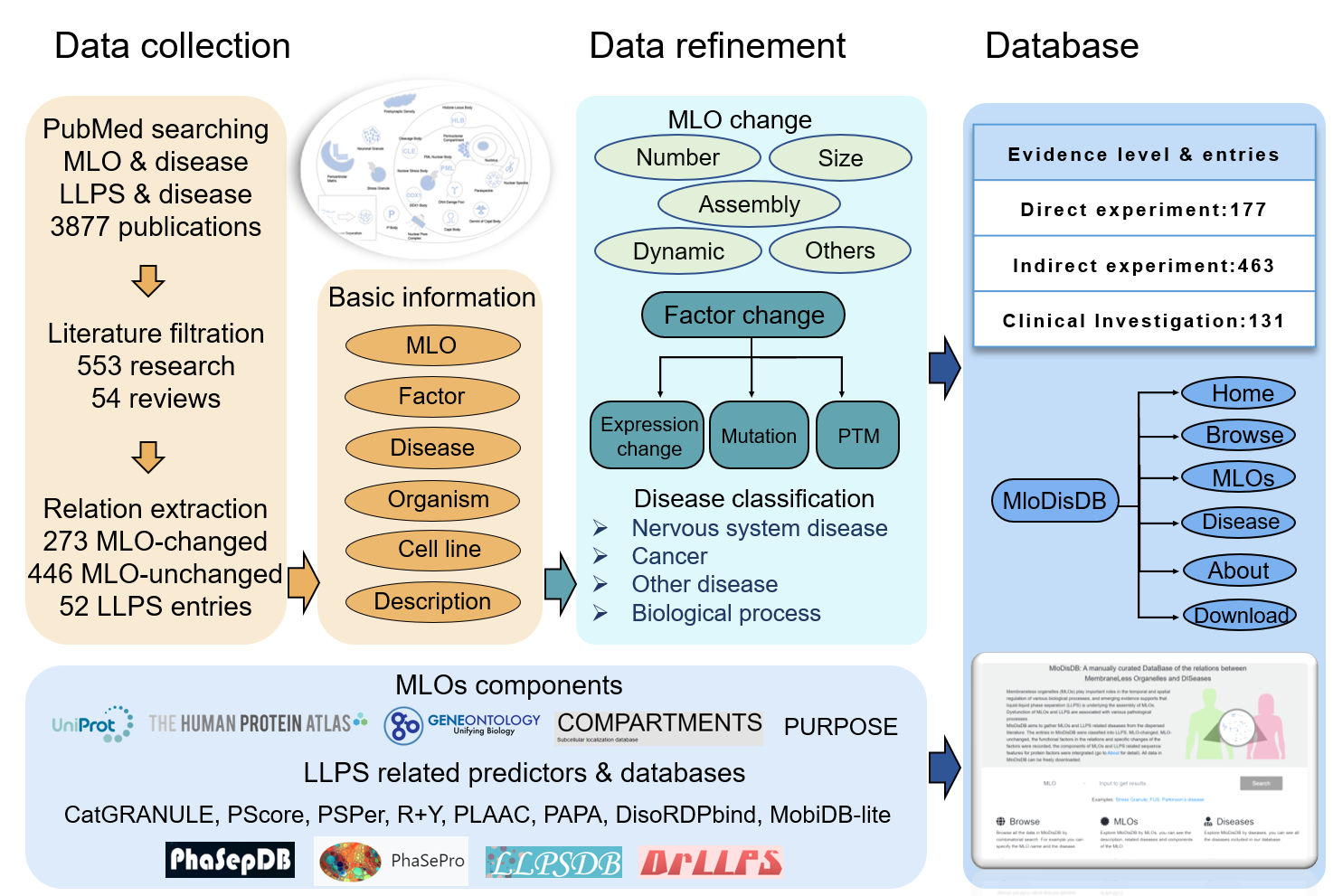
2. Data collection
3877 Publications were obtained by searching NCBI PubMed using keyword
including MLOs and diseases. For example, the search keyword for stress granule
is
“((stress granules[title/abstract]) or (stress granule[title/abstract])) and ((disease[title/abstract]) or (cancer[title/abstract]) or (neurodegeneration [title/abstract]))”
Detailed keywords list can be found in the Download page.
All the publications in MloDisDB were published before 2020/04/01. 29
MLOs were searched and 15 of them had related publications which met our
criteria. Relations between LLPS and diseases were searched separately and displayed in parallel
with MLOs in MloDisDB.
As of June 2020, we have manually filtered 607 publications (553 research papers and 54 reviews) that describe 719 relationships between MLOs and diseases and 52 relationships between LLPS and diseases. Related information was extracted from filtered publications, such as the organisms and cell lines used for experiments, descriptions of the relations, the factors that function in the process etc.

3. Entry Classification
The entries were classified into three classes: LLPS, MLO-changed and MLO-unchanged. LLPS entries described LLPS-disease relations. MLO-changed and MLO-unchanged entries described MLO-disease relations, they were classified based on whether the MLOs changed or not in the relations. The size, number, assembly and dynamic changes of MLOs and the descriptions of the changes were extracted for MLO-changed entries.

4. Experiment Evidence
Each entry was assigned with one of the three evidence levels based on original publication.
1) “Direct experiment”: the abnormal of the MLO or factor causes typical symptoms of the disease in model organisms or cell lines.
2) “Indirect experiment”: the abnormal of the MLO or factor brings about certain changes which are indicative of the development of the disease; or the original publication focuses on a known disease-causing factor, and the disease related changes of the factor perturb the MLO.
3) “Clinical Investigation”: the relation is extracted from clinical samples investigation or drug usage investigation.
5. Disease Classification
The diseases were mapped to Disease Ontology database, Online Mendelian Inheritance in Man (OMIM), Medical Subject Headings (MeSH) and ICD-10 Clinical Modification, and were classified into four categories:
1) Nervous system diseases,
2) Cancers,
3) Other diseases like infectious diseases and anemia,
4) Biological processes like apoptosis and aging.
The last category means that the MLO is important in the biological process but the original
publication didn't show direct link with disease.
6. Factors
The functional factors in the MLO-disease relations and LLPS-disease relations were classified into three categories: protein, RNA and others (such as chemicals and artificially synthesized oligonucleotides, peptides). Some entries described that the perturbations of the MLOs were related to the diseases, and no factor was described in the original publication, “None” was recorded for these entries. Some studies further presented the specific changes on the factor which contributing to the diseases, therefore, 247 expression changes, 199 mutations and 46 post-translational modifications were extracted and recorded.


7. Components collection
The components of MLOs were obtained from 5 resources: UniProt, Gene Ontology, The Human Protein Atlas, COMPARTMENTS, Protein Universal Reference Publication-Originated Search Engine (PURPOSE). COMPARTMENT and PURPOSE are text mining based tools. Known LLPS proteins were collected from the reviewed proteins of PhaSepDB with in vitro experiment evidence, the reviewed scaffolds of DrLLPS, the proteins in PhaSePro, and the natural proteins of LLPSDB. Only reviewed human proteins were included in MloDisDB.
8. LLPS related predictions
The LLPS related predictions were predicted via CatGRANULE, PScore, PSPer, R+Y, PLAAC and PAPA. DisoRDPbind and MobiDB-lite which
provide intrinsic disorder region related predictions were included as well. the links of the predictors were listed below.
In MloDisDB, R+Y was calculated as (N_R*N_Y)/6500. N_R, N_Y represent the number of arginine and tyrosine residues in IDRs, IDRs are defined as the regions whose MoBiDB-lite >= 3/8.
9. Related links
| Disease Ontology | The Disease Ontology has been developed as a standardized ontology for human disease with the purpose of providing the biomedical community with consistent, reusable and sustainable descriptions of human disease terms. |
|---|---|
| Online Mendelian Inheritance in Man (OMIM) | An Online Catalog of Human Genes and Genetic Disorders. |
| Medical Subject Headings (MeSH) | The Medical Subject Headings (MeSH) thesaurus is a controlled and hierarchically-organized vocabulary produced by the National Library of Medicine. It is used for indexing, cataloging, and searching of biomedical and health-related information. MeSH includes the subject headings appearing in MEDLINE/PubMed, the NLM Catalog, and other NLM databases. |
| ICD-10 Clinical Modification | International Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM). |
| PhaSepDB | The database of phase-separation related proteins |
| PhaSePro | PhaSePro is the comprehensive database of proteins driving liquid-liquid phase separation (LLPS) in living cells |
| DrLLPS | Data resource of liquid-liquid phase separation |
| LLPSDB | A database of proteins undergoing liquid-liquid phase separation in vitro |
| UniProt | The mission of UniProt is to provide the scientific community with a comprehensive, high-quality and freely accessible resource of protein sequence and functional information. |
| AmiGO 2 | Gene Ontology. |
| The Human Protein Atlas | The Human Protein Atlas is a Swedish-based program initiated in 2003 with the aim to map all the human proteins in cells, tissues and organs using integration of various omics technologies. |
| PURPOSE | Systematic Proteins Prioritization in Organ Systems and Diseases through Literature Mining. |
| COMPARTMENTS | COMPARTMENTS is a weekly updated web resource that integrates evidence on protein subcellular localization from manually curated literature, high-throughput screens, automatic text mining, and sequence-based prediction methods. |
| PLAAC | PLAAC searches protein sequences to identify probable prion subsequences using a hidden-Markov model (HMM) algorithm. |
| PSPer | Unsupervised prediction of proteins able to form phase-separated liquid droplets acting as membraneless organelles. |
| Pscore | Pi-Pi contacts are an overlooked protein feature relevant to phase separation. |
| catGRANULE | catGRANULE is an algorithm to predict liquid-liquid phase separation propensity (LLPS). |
| R+Y | Critical concentration prediction based on number of arginine and tyrosine residues, extrapolated from FET family proteins. |
| PAPA | a method for predicting prion forming propensity. |
| MobiDB | a database of protein disorder and mobility annotations. |
| DisoRDPbind | DisoRDPbind predicts the RNA-, DNA-, and protein-binding residues located in the intrinsically disordered regions. |
Chao Hou, Haotai Xie, Yang Fu, Yao Ma, Tingting Li. MloDisDB: A manually curated DataBase of the relations between MembraneLess Organelles and DISeases